VISHC researchers Cuong Do and Hieu Pham, together with two VinUni undergrad students have developed a new deep-learning system to detect sleep apnea accurately from ECG signals. The new method has higher performance compared to existing methods. This new study just has been accepted for publication in the IEEE Journal of Biomedical and Health Informatics (J-BHI).

Xuan Hieu and Viet Duong (right) – undergraduate students at VinUni’s College of Engineering and Computer Science present their research at the VinUni-Illinois Smart Health Center.
Sleep apnea (SA) is a common sleep disorder characterised by brief interruptions of breathing during sleep. These episodes usually last 10 seconds or more and occur repeatedly throughout the night. The most common type of sleep apnoea is obstructive sleep apnoea (OSA) caused by relaxation of soft tissue in the back of the throat that blocks the passage of air. When a person has sleep apnea, their breathing repeatedly stops and starts during sleep, which can result in low levels of oxygen. Symptoms include daytime sleepiness, loud snoring, restless sleep, and more. Sleep apnea (SA) is a significant respiratory condition that poses a major global health challenge.
According to The National Council of Aging, roughly 6 million Americans have been diagnosed with sleep apnea. But in reality, this sleep disorder is thought to affect 30 million people in the U.S. Older adults are much more likely to have sleep apnea than younger people. One study found that 56% of people age 65 and older have a high risk of developing obstructive sleep apnea. Not all people with sleep apnea have an official diagnosis. The American Academy of Sleep Medicine (AASM) estimates that as many as 80% of people with OSA are undiagnosed.
In this study, VISHC researchers proposed a deep learning-based approach to address this diagnostic gap by delving deeper into the comprehensive segments of the ECG signal. The proposed methodology draws inspiration from Matrix Profile algorithms, which generate an Euclidean distance profile from fixed-length signal subsequences. From this, the authors derived the Min Distance Profile (MinDP), Max Distance Profile (MaxDP), and Mean Distance Profile (MeanDP) based on the minimum, maximum, and mean of the profile distances, respectively. Their technique, called MPCNN demonstrated a significant potential and efficacy for SA classification, delivering promising per-segment and per-recording performance metrics.
“We are really excited about the proposed approach, which goes beyond conventional ECG signal analysis by considering distance relationships within comprehensive signal segments, thereby improving the accuracy of SA detection” said authors Xuan Hieu and Viet Duong.
The proposed approach achieves per-segment accuracy of up to 92.11% and per-recording accuracy of 100%, outperforming state-of-the-art SA detection methods.The method’s robustness is demonstrated through testing with lightweight models such as modified LeNet-5, BAFNet, and SEMSCNN.
“We believe that this study opens avenues for practical applications, including HSAT and SA detection in Internet of Things (IoT) devices, contributing to the advancement of accessible and effective healthcare solutions” – Assistant Professors Cuong Do and Hieu Pham – Researchers at the VinUni-Iillinois Smart Health Center added.
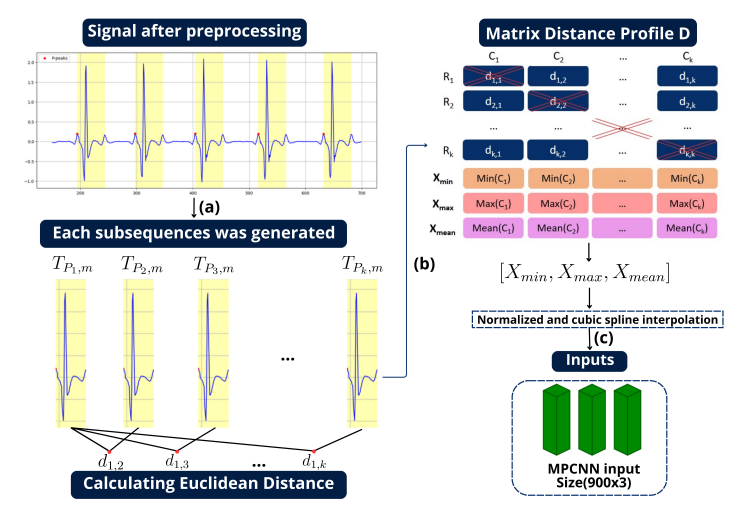
Detailed procedure of the MPCNN. In the preprocessing stage, subsequences are generated, represented by the yellow window in the image. Each subsequence begins at a P peak and extends for a window size of m. The Matrix Distance Profile is created based on the Euclidean Distance. Subsequently, important features are calculated based on the minimum, maximum, and mean values from each column of the matrix. The input is then normalized to the range [0, 1] and subjected to cubic spline interpolation to achieve a final dimension.
—————
Editor’s note:
The paper titled “MPCNN: A Novel Matrix Profile Approach for CNN-based Sleep Apnea Classification” by Hieu X. Nguyen, Duong V. Nguyen, Hieu H. Pham, and Cuong D. Do can be accessed in its full version on ArXiv. Its published version can be found by Digital Object Identifier or DOI: 10.1109/JBHI.2024.3397653











